About the Laboratory of Microbial Enzyme Potential (Onaka Lab.)
The Laboratory of Microbial Enzyme Potential (Amano Enzyme) is a laboratory endowed by Amano Enzyme Inc. at the Graduate School of Agricultural and Life Sciences, the University of Tokyo, since 2017.
“Microorganism” is a generic term for tiny invisible organisms. These ubiquitous organisms are soil decomposers, and are involved in maintaining global environment such as the carbon and nitrogen cycles. In addition, they are normal inhabitants in the human body, pathogens for mammals and plants, and producers of a wide variety of organic materials. However, usually, we do not mind their presence in our daily life.
The recent advances in microbial genome analysis have changed the perception of microorganisms. Microbial genomic information revealed that microorganisms have biological diversity far beyond that between mammals and plants, and we are expecting to discover novel microbial functions from these genomic data.
In the Laboratory of Microbial Enzyme Potential, we are exploring the hidden gifts and talents of the microorganisms and applying them in industrial processes. In particular, we would be focusing on actinomycetes, soil bacteria known for their ability to produce antibiotics, and on identifying novel potent biosynthetic machinery in their secondary metabolism.
The current research interests of our laboratory are (i) development of technology to promote antibiotic production capacity of actinomycetes, (ii) biosynthetic studies of RiPPs for drug discovery based on medium-sized molecules, (iii) studies regarding fermented foods and beverages: conventional fermented foods and their relationship to new fermentation methods.

Our Research

Development of technology to promote antibiotic production capacity of actinomycetes
The development of new drugs relies markedly on the discovery of novel natural products that are produced by microorganisms. The soil microorganisms, actinomycetes, are known to produce various secondary metabolites. The recent genomic analyses of some Streptomyces strains have revealed the presence of biosynthetic gene clusters for approximately 30 secondary metabolites, implying that a single Streptomyces strain can produce over 30 secondary metabolites. However, some of these secondary metabolite genes are cryptic in fermentation cultures, and because many biosynthetic genes for secondary metabolites remain silent under typical laboratory culture conditions, the valuable secondary metabolites which they produce, have not been investigated. We have therefore developed new methods to screen for the secondary metabolites.
(1) Combined-culture, a new co-culture method for screening of natural products
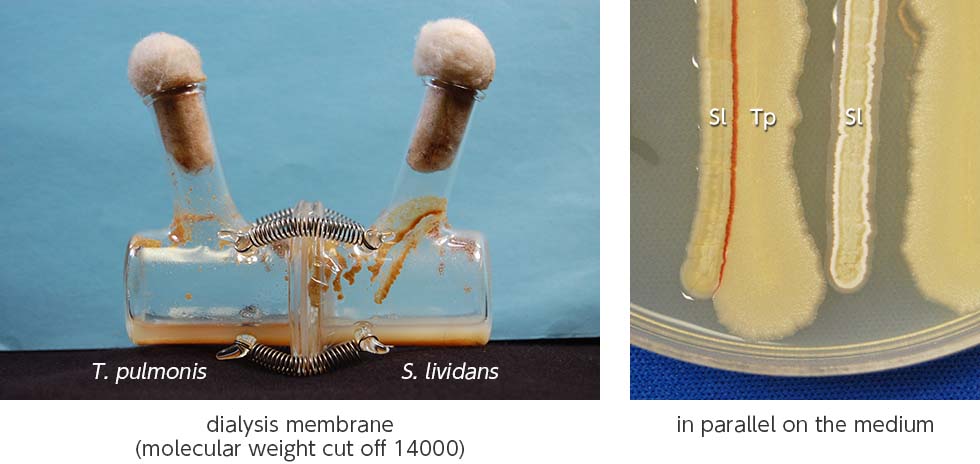
We discovered that the mycolic acid localized in the outer cell layer of an inducer bacterium affects the secondary metabolism in Streptomyces, and is due to the physical contact between the mycolic acid-containing bacteria (MACB: Mycolic Acid Containing Bacteria) and Streptomyces. As an example, the production of red pigment by Streptomyces lividans TK23 was induced during its co-culture with Tsukamurella pulmonis TP-B0596 which contains mycolic acid; however, no pigment was observed when the two bacteria were partitioned (Fig. 1-1).
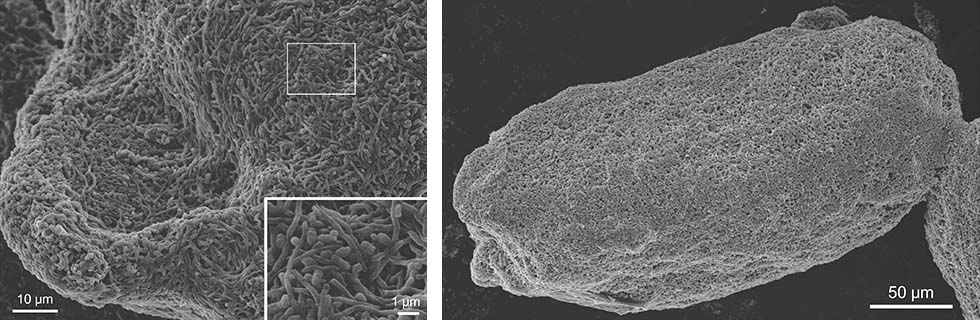
Observations of the combined-culture via scanning electron microscopy (SEM) indicated that the adhesion of live MACB to S. lividans mycelia was a significant interaction that resulted in co-aggregations (Fig. 1-2).
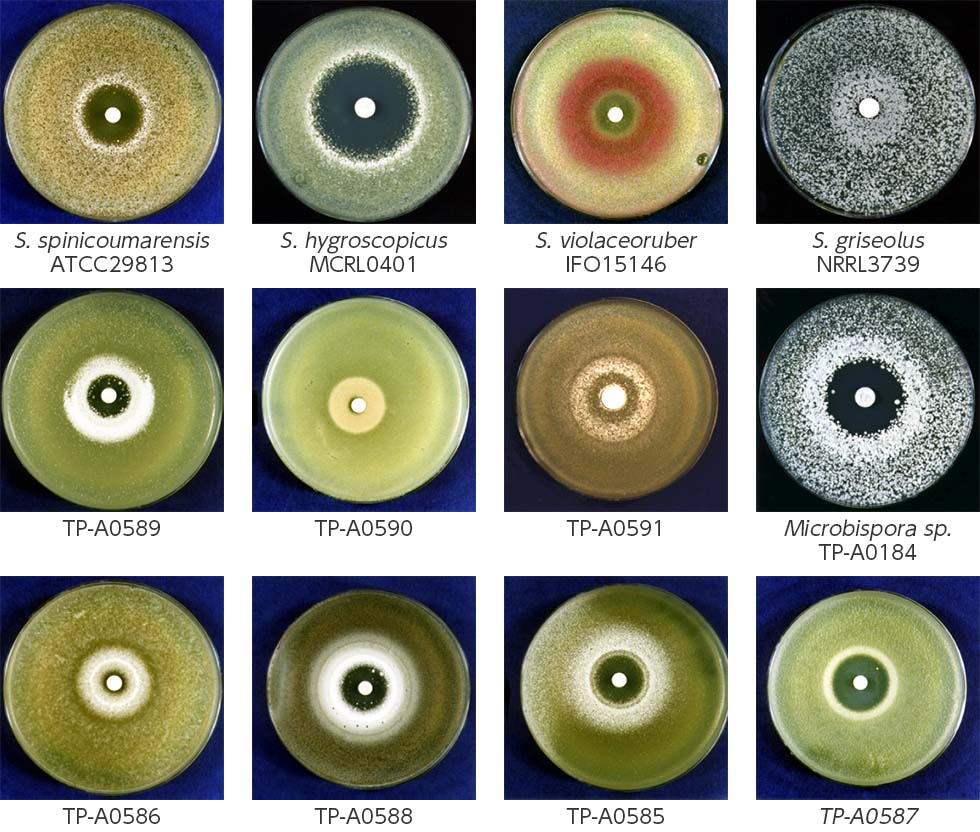
We developed a new co-culture method known as the “combined-culture” method based on these results. The combined culture method facilitates the screening of natural products. To date, we have discovered more than 40 novel compounds using the combined culture method (Figs. 1-3 and 1-4).
Key reference papers pertaining to combined culture
- The often cited paper by Onaka H, Mori Y, Igarashi Y, & Furumai T. (Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl Environ Microbiol 77(2): 400-406 (2011). doi: 10.1128/AEM.01337-10) was the first to report the phenomenon of combined-culture.
- The paper by Onaka H, Ozaki T, Mori Y, Izawa M, Hayashi S, & Asamizu S. (Mycolic acid-containing bacteria activate heterologous secondary metabolite expression in Streptomyces lividans. J Antibiot (Tokyo) 2015 Apr 1. doi: 10.1038/ja.2015.31) mentions that the combined-culture method is effective for heterologous expression.
- This study performed by Asamizu S, Ozaki T, Teramoto K, Satoh K, & Onaka H. (Killing of mycolic acid-containing bacteria aborted induction of antibiotic production by Streptomyces in combined-culture. PLoS One. 2015 Nov 6;10(11):e0142372. doi: 10.1371/journal.pone.0142372) discovered that the co-aggregation by physical contact between actinomycetes and MACB is important for the induction of secondary metabolism in combined-cultures.
- In this study, the authors, M. Kato, S. Asamizu, & H. Onaka (Intimate relationships among soil bacteria: actinomycetes and mycolic acid-containing bacteria. Scientific Reports, (in press)) suggest that pairs of actinomycetes and MACB co-aggregate and live together in natural environments.
- The authors, S. Hoshino, H. Onaka, & I. Abe compiled a review covering the secondary metabolites induced by combined-cultures. (Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes co-cultured with mycolic acid-containing bacteria. J Indust Microbiol Biotechnol 46(3-4):363-374 (2019).).
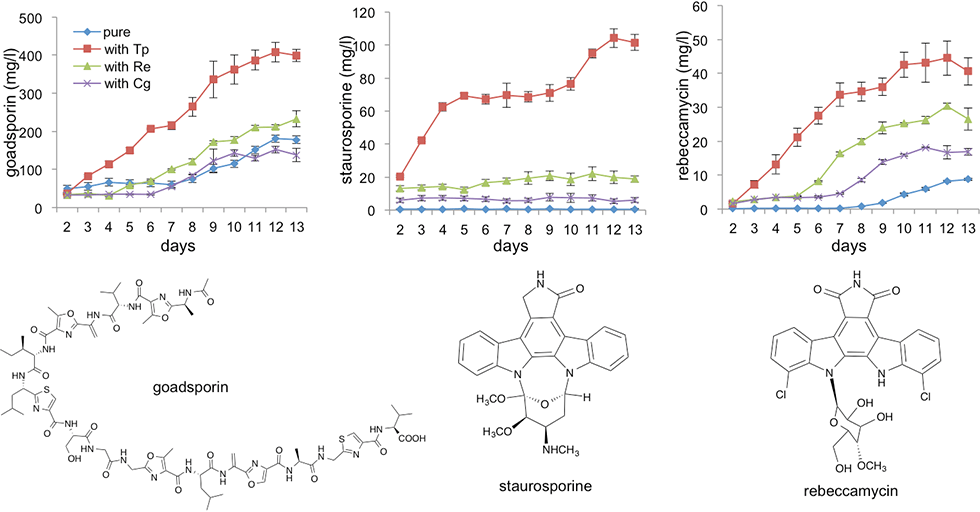
(2) Goadsporin (GS): a natural product that induces secondary metabolism in various actinomycetes
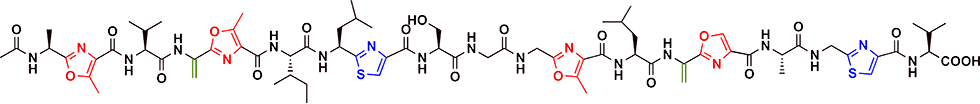
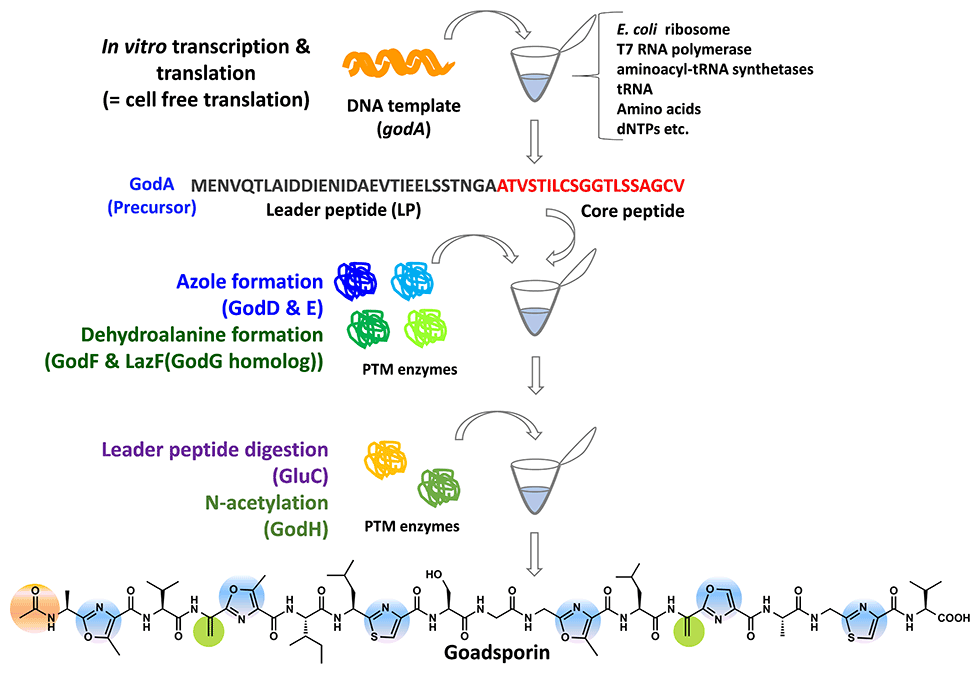
GS is a linear polypeptide antibiotic produced by the Streptomyces sp. TP-A0584 (Fig. 2-1). GS comprises 19 amino acids, including eight unusual amino acids, and oxazole, thiazole, and dehydroalanine. GS promotes the production of secondary metabolites, and morphogenesis at low concentrations and induces growth inhibition at high concentrations in actinomycetes (Fig. 2-2).
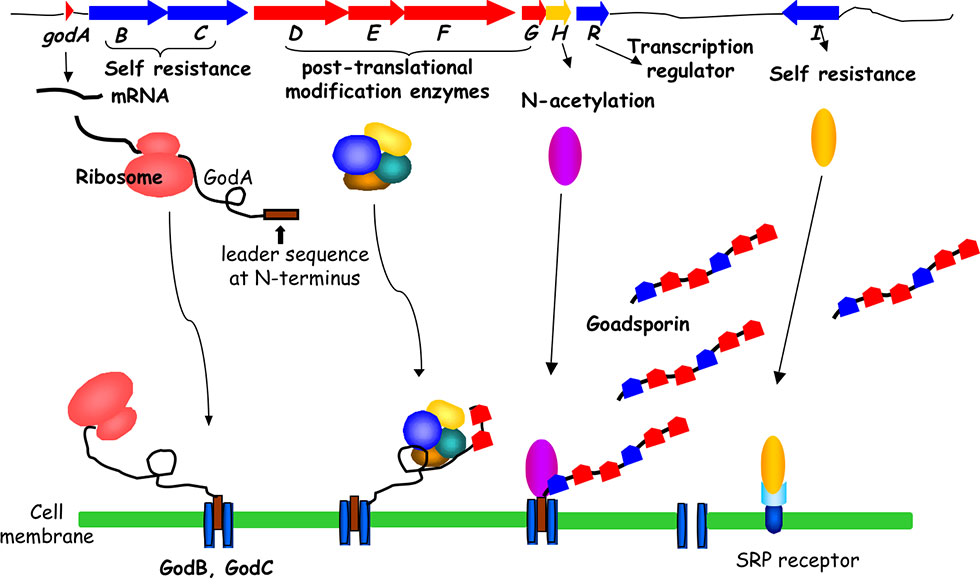
The GS biosynthetic gene cluster contains a structural gene, godA and nine god (GS) genes that participate in post-translational modification, immunity, and transcriptional regulation, spanning 20 kb. GS biosynthesis is initiated by the translation of 49 amino acid GodA polypeptides. The subsequent dehydration to form dehydroalanine is catalyzed by GodF and GodG, while the cyclization to form oxazole and thiazole rings is catalyzed by GodD and GodF. Finally, the N-terminal leader sequence of 30 amino acids is digested and acetylated by GodH acetyltransferase to achieve GS (Fig. 2-3).
Key reference papers pertaining to GS
- The paper by H. Onaka, H. Tabata, Y. Igarashi, Y. Sato, & T. Furumai (Goadsporin, a chemical substance which promotes secondary metabolism and morphogenesis in streptomycetes. I. Purification and Characterization. J Antibiot 54:1036-1044 (2001). doi: 10.7164/antibiotics.54.1036) mentions the identification of GS and its bioactivity.
- The study by Onaka H, Nakaho M, Hayashi K, Igarashi Y, & Furumai T. (Cloning and characterization of goadsporin biosynthetic gene cluster from Streptomyces sp. TP-A0584. Microbiol 151: 3923-3933 (2005). doi: 10.1099/mic.0.28420-0) discovered that GS is a ribosomally synthesized and post-translationally modified peptide (RiPP).
- This paper by Ozaki T, Kurokawa Y, Hayashi S, Oku N, Asamizu S, Igarashi Y, & Onaka H (Insights into the biosynthesis of dehydroalanines in goadsporin. Chem Bio Chem (2015). doi: 10.1002/cbic.201500541) mentions the mechanism by which dehydroalanine is involved in GS biosynthesis.

Biosynthetic studies of RiPPs for drug discovery based on medium-sized molecules
Medium-sized molecules with molecular weights between 500 and 2,000 benefit drug discovery as they exhibit substrate specificities comparable with those of biopharmaceuticals, such as antibodies, and can be produced at a low cost, similar to small molecules. Among the medium-sized molecules, peptide-based natural products have received attention because they can be produced via fermentation. Fermentation is more cost effective than the production of synthetic peptides.
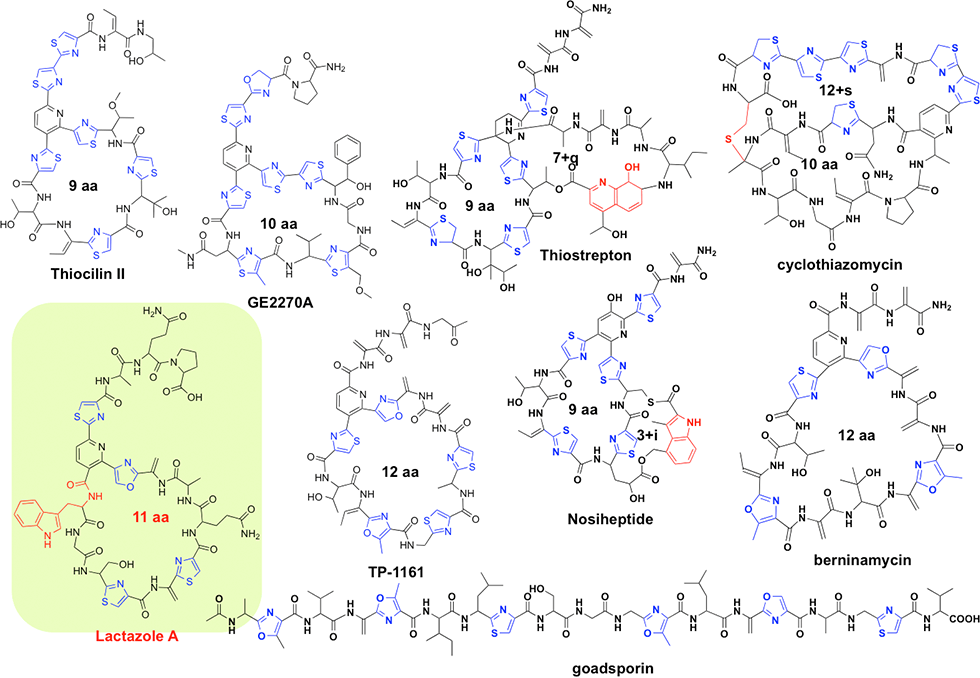
We focused on RiPPs, which is a group of peptide-based compounds in which the amino acids are modified by post-translationally modified enzymes after ribosome synthesis. RiPPs undergo post-translational modifications, therefore, they are more likely to form a stable three-dimensional structure and exhibit a higher affinity for the target as compared with non-modified peptides. We have investigated the biosynthesis of GS, lactazoles, and goadvionins, and have synthesized hundreds of RiPP analogues exchanged the amino acid sequences (Fig. 4-2). RiPPs are stable in living cells because they are less likely to be degraded by peptidases and other enzymes.
(1) Lactazole: thiopeptide produced via minimum biosynthetic gene set
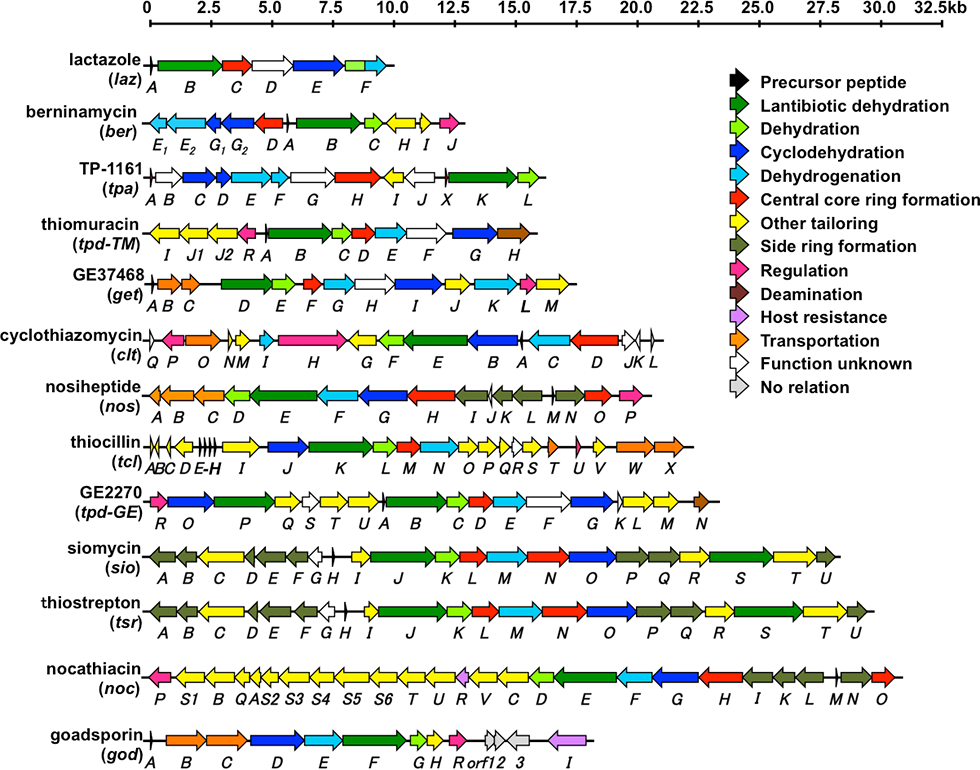
Thiopeptides are produced primarily by actinomycetes and typically contain highly modified sulfur-containing peptides; the peptides feature a characteristic macrocycle knotted with pyridine or piperidine; pyridine and piperidine are a six-membered nitrogen-containing ring structures. Thiopeptides are a group of antibiotics that, similar to GS, are biosynthesized via the ribosomal translation system. More than 100 thiopeptides have been identified to date. We performed genome mining to identify a cryptic thiopeptide biosynthetic gene cluster that participates in the biosynthesis of lactazoles from Streptomyces lactacystinaeus OM-6519 (Fig. 3-1).
Lactazoles originate from the simplest cluster, which contains only six unidirectional genes (lazA to lazF). This cluster is the smallest cluster among the known thiopeptide biosynthetic gene clusters (Fig. 3-2). The structure gene, lazA, contains the precursor peptide sequence and is classified into a phylogenetically distinct clade. lazC participates in the macrocyclization process and aids the formation of a central pyridine moiety via gene disruption. This compact biosynthetic machinery is expected to exhibit high potency, which will result in diverse thiopeptide core structures. Our approach facilitates the production of a more diverse set of thiopeptide structures, thereby increasing the range of semi synthetic RiPPs for use in drug development.
Key reference paper pertaining to lactazole biosynthesis
- This paper by S. Hayashi, T. Ozaki, S. Asamizu, H. Ikeda, S. Ōmura, N. Oku, Y. Igarashi, H. Tomoda, & H. Onaka (Genome mining reveals a minimum gene set for the biosynthesis of 32-membered macrocyclic thiopeptides lactazoles. Chem Biol 21(5):679-88 (2014). https://doi.org/10.1016/j.chembiol.2014.03.008) mentions the identification of lactazoles via genome mining.
(2) Development of technology to extend structural diversity of peptide antibiotics
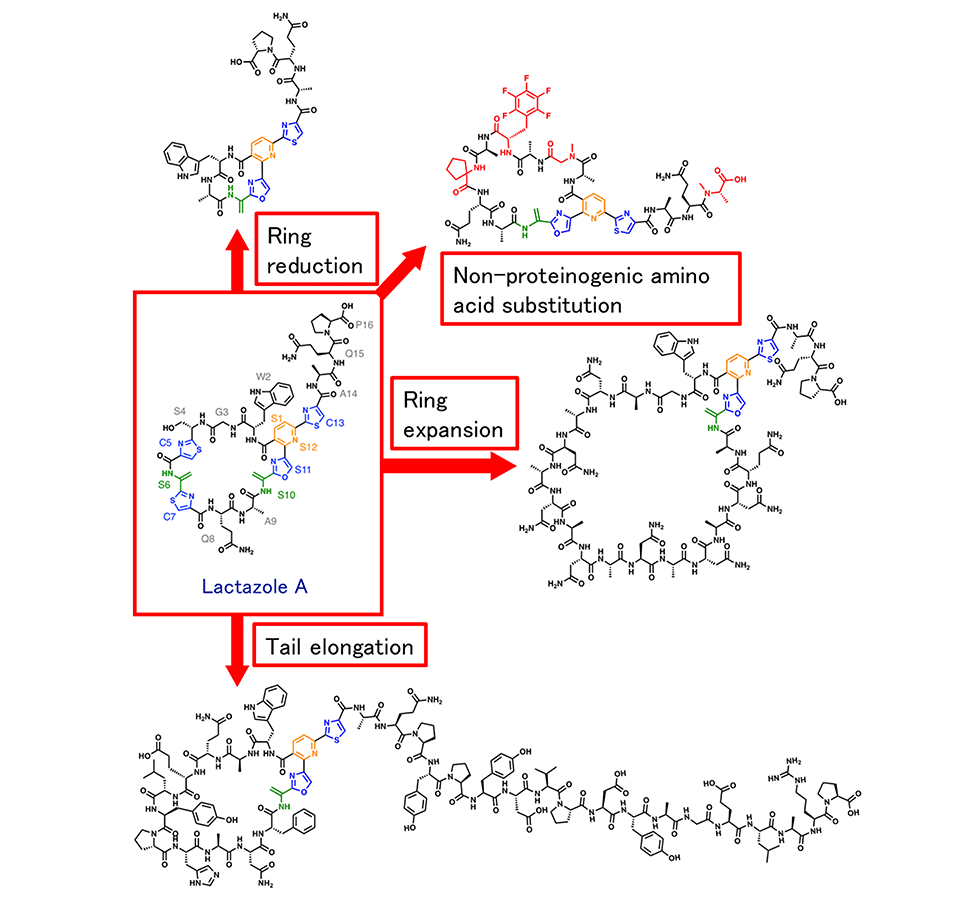
We are the first group in the world to successfully reconstitute an entire RiPP biosynthetic pathway using an in vitro one-pot reaction. The system comprises a cell-free translation system and an in vitro post-translational modification system, in which the precursor peptide gene is first introduced into the reaction tube, and GS, lactazole, and their analogues are synthesized overnight (Fig. 4-1). This epoch-generating system can rapidly and easily synthesize various RiPP analogues containing different amino acid sequences in one day.
As for the synthesis of lactazole analogues, since the substrate recognition of the macrocyclophilic enzyme LazC was found to be permissive, we successfully created analogues comprising various ring sizes, and analogues comprising amino acid sequences within the ring; the analogues differed completely from the original. Additionally, we successfully introduced non-proteinaceous amino acids by using the cell-free translation system (Fig. 4-2).
Key reference papers pertaining to syntheses of various RiPPs analogues
- In this paper, (Minimal lactazole scaffold for in vitro thiopeptide bioengineering. Nature Comm 11, 2272 (2020). https://doi.org/10.1038/s41467-020-16145-4), the authors, A. A. Vinogradov, M. Shimomura, Y. Goto, T. Ozaki, S. Asamizu, Y. Sugai, H. Suga, & H. Onaka report the in vitro one-pot reaction of lactazole biosynthesis and the synthesis of diverse lactazole analogues.
- The paper written by T. Ozaki, K. Yamashita, Y. Goto, M. Shimomura, S. Hayashi, S. Asamizu, Y. Sugai, H. Ikeda, H. Suga, & H. Onaka, (Dissection of goadsporin biosynthesis by in vitro reconstitution leads to designer analogues expressed in vivo. Nature Comm e14207 (2017). https://doi.org/10.1038/ncomms14207) reports the in vitro one-pot reaction of GS biosynthesis and the syntheses of 52 GS analogues.
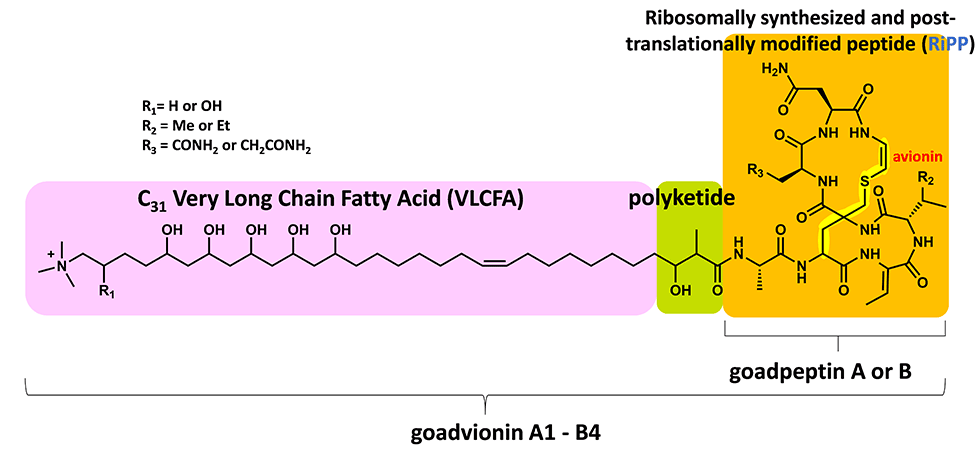
(3) Novel lipopeptide goadvionins
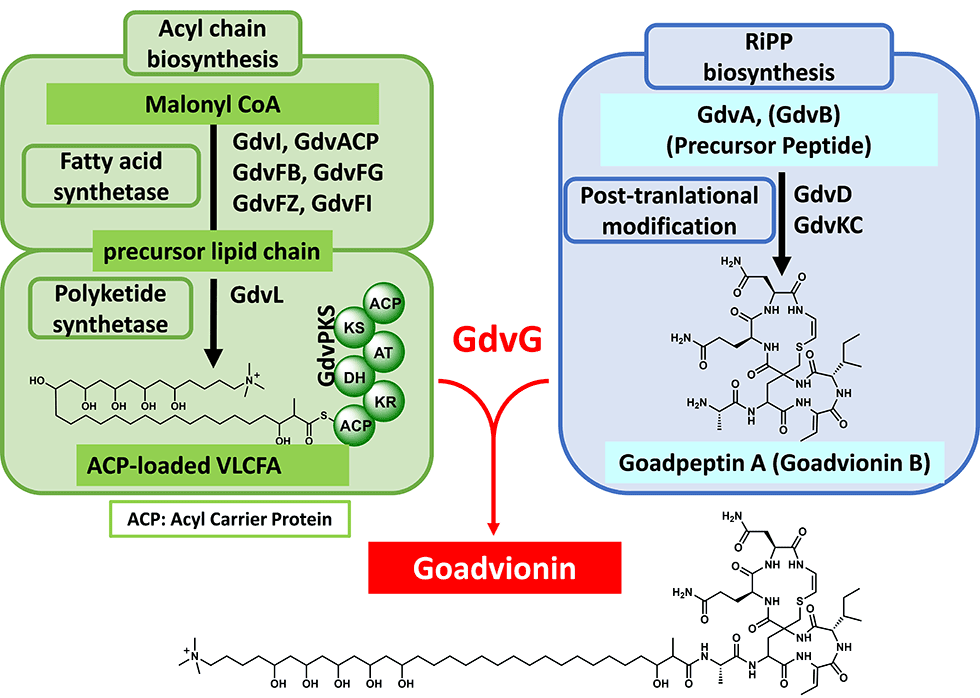
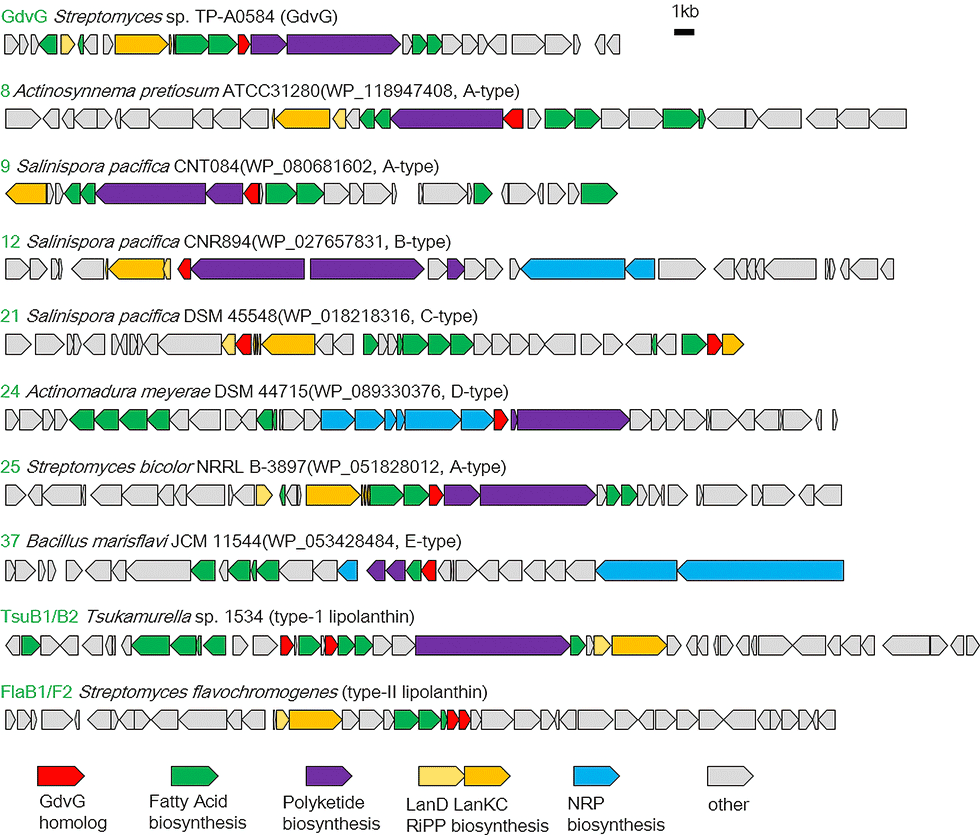
Goadvionins are hybrid polyketide (PK)/RiPP hybrid compounds synthesized via a biosynthetic pathway, which differs from the typical lipopeptide biosynthetic pathway (Fig. 5-1). Whereas most lipopeptides are composed of non-ribosomal ribosomal synthase (NRPS), goadvionins are biosynthesized via the condensation of RiPPs and PK by a novel acyltransferase, GdvG (Fig. 5-2).
Key reference paper pertaining to goadvionin
- The paper (Acyltransferase that catalyses the condensation of polyketide and peptide moieties of goadvionin hybrid lipopeptides. Nature Chem 12, 869–877 (2020). doi: 10.1038/s41557-020-0508-2 ) by R. Kozakai, T. Ono, S. Hoshino, H. Takahashi, Y. Katsuyama, Y. Sugai, T. Ozaki, K. Teramoto, K.Teramoto, K. Tanaka, I. Abe, S. Asamizu, & H. Onaka, reports the isolation, structural analysis, biosynthesis, and bioactivity of goadvionins.

Studies regarding fermented foods and beverages: conventional fermented foods and their relationship to new fermentation methods
(1) Development of Alcoholic Beverages Using Wild Yeasts
Professor Onaka isolated wild sake brewing yeast from barley malt in Toyama and assisted in the development of Toyama-brewed craft beer as part of the university’s regional collaboration activities. “Yeast born in Toyama” (Fig. 6-1) exhibits a high alcohol fermentation ability and has been used for brewing Toyama-brewed craft beer, and for brewing Japanese sake, wine, and whiskey.
In addition, ccPTM18, a wild yeast isolated from wine fermenting in barrels at COCOFARM & WINERY (Ashikaga, Japan), is characterized by the acidity it yields. When it is used in Japanese sake brewing, it produces sake that is similar to white wine and has a pleasant floral note and a sour taste.
Each year, sake is produced using these wild yeasts and is enjoyed by many people (Fig. 6-2).


(2) Fermented Foods Worldwide
Various fermentation cultures exist worldwide. Traditional fermentation is used primarily to preserve fresh foods for long durations; however, recently, the benefits of consuming of fermented foods, have been prioritized. By independently creating various fermented foods from countries worldwide, ourselves, we can obtain a deeper understanding of fermented foods, which will promote the development of new fermented foods.
We welcome joint research and consultation with companies, stores, etc.
Click to read an article that describes each fermented food. It is available in Japanese only, however, many pictures are included and will aid your understanding.
Address
Laboratory of Microbial Metabolic Potential
Department of Biotechnology, Graduate School of Agricultural and Life Sciences,
The University of Tokyo.
1-1-1 Yayoi, Bunkyo, Tokyo 113-8657, Japan.








Current members

Hiroyasu Onaka
Specially-appointed Professor (Ph.D.)
Education
- The University of Tokyo, Tokyo, Japan A.B. 1993
- The University of Tokyo, Tokyo, Japan Ph.D. 1998
- Postdoctoral Fellow at Cold Spring Harbor lab., NY, U.S.A. & Penn State University, PA, U.S.A. 1998-1999
Professional Experience
- 1997-1999 JSPS (Japan Society for the Promotion of Science) Research Fellowship for Young Scientist
- 1999-2006 Assistant professor at the Department of Biotechnology, Toyama Prefectural University, Toyama, Japan.
- 2006-2010 Lecturer at the Department of Biotechnology, Toyama Prefectural University, Toyama, Japan.
- 2010-2012, Associate Professor at the Department of Biotechnology, Toyama Prefectural University, Toyama, Japan.
- 2012- Specially-appointed professor of Graduate school of Agricultural and Life Sciences, The University of Tokyo
Society Memberships and Awards
- Hamada Award, the Society for Actinomycetes Japan, 2005
- The Japan Bioscience, Biotechnology and Agrochemistry Society Award for the Encouragement of Young Scientists, Japan society for Bioscience, Biotechnology and Agrochemistry, 2008
- Toyama Award, Toyama Prefecture Citizens' Personal Development Foundation, 2010
- Specially appointed Fellow of Toyama Prefectural University, 2012-
- The Sumiki-Umezawa Memorial Award, The Japan Antibiotics Research Association, 2016
- Omura Award, the Society for Actinomycetes Japan, 2022

Shumpei Asamizu
Specially-appointed Lecturer (Ph.D.)
Education
- 2001.4-2005.3 Toyama Prefectural University (Bachelor of Agriculture from NIAD-UE)
- 2005.4-2007.3 Toyama Prefectural University (Master of Engineering)
- 2007.4-2010.3 Toyama Prefectural University (Doctor of Engineering)
Professional Experience
- 2008.4-2010.3 JSPS Research Fellow (DC2)
- 2010.4-2013.3 Oregon State University, Postdoctoral Research Associate (Taifo Mahmud Lab.)
- 2013.4-2019-4 Specially-appointed Assistant Professor of Graduate school of Agricultural and Life Sciences, The University of Tokyo
- 2019.5- Specially-appointed Lecturer of Graduate school of Agricultural and Life Sciences, The University of Tokyo
Society Memberships and Awards
- Hamada Award, the Society for Actinomycetes Japan, 2017
- The Japan Bioscience, Biotechnology and Agrochemistry Society Award for the Encouragement of Young Scientists, Japan society for Bioscience, Biotechnology and Agrochemistry, 2017
Ph.D. Thesis
Studies on Bis-indole Natural Products Biosynthesis from Bacteria (Toyama Prefectural University, 2010.3)
Interests, Keywords
Secondary metabolism, Biosynthesis, Enzyme chemistry, Secondary metabolism gene regulation, Natural products chemistry, Synthetic biology, Metabolic engineering, Protein Engineering

Shotaro Hoshino
Specially-appointed Assistant Professor (Ph.D.)
Education
- 2010.4 - 2014.3 The University of Tokyo (Bachelor of Pharmaceutical Science)
- 2014.4 - 2016.3 The University of Tokyo (Master of Pharmaceutical Science)
- 2016.4 - 2019.3 The University of Tokyo (Doctor of Pharmaceutical Science)
Professional Experience
- 2016.4 - 2019.3 JSPS Research Fellow (DC1)
- 2019.6 - 2021.11 JSPS Overseas Research Fellowships
- 2019.6 - 2022.2 Princeton University, Department of Chemistry, Postdoctoral Researcher (Prof. Mohammad R. Seyedsayamdost Lab.)
- 2022.4 - Specially-appointed Assistant Professor of Graduate school of Agricultural and Life Sciences, The University of Tokyo
Ph.D. Thesis
Searching for rare-actinomycetes-derived novel drug seeds by combined-culture method (The University of Tokyo, 2019.3)
Interests, Keywords
Natural product chemistry, Isolation & structural elucidation, microbial interaction, Natural product biosynthesis, Regulation of secondary metabolism, Enzymology, Actinobacteria

Emiko Nagai
Postdoctoral Fellow

Manami Kato
Student in the 4th year of the doctoral program

Yukun Lei
Student in the 3rd year of the doctoral program

Shinta Ijichi
Student in the 2nd year of the master's program
Publications
2022
- A. A. Vinogradov, J. S. Chang, H. Onaka, Y. Goto, and H. Suga. Accurate Models of Substrate Preferences of Post-Translational Modification Enzymes from a Combination of mRNA Display and Deep Learning. ACS Central Science, (in press)
- M. Kato, S. Asamizu, and H. Onaka*. Intimate relationships among soil bacteria: actinomycetes and mycolic acid-containing bacteria. Scientific Reports 12: 7222 (2022)
- Y. Asai, T. Hiratsuka, M. Ueda, Y. Kawamura, S. Asamizu, H. Onaka, M. Arioka, S. Nishimura, M. Yoshida*. Differential Biosynthesis and Roles of Two Ferrichrome-Type Siderophores, ASP2397/AS2488053 and Ferricrocin, in Acremonium persicinum. ACS Chemical Biology, 17(1), 207-216 (2022)
- A. Hikima, S. Asamizu, H. Onaka, H. Zhang, H. Tomoda*, N. Koyama. Kimidinomycin, a New Antibiotic against Mycobacterium avium Complex, Produced by Streptomyces sp. KKTA-0263. The Journal of Antibiotics, 75(2), 72-76 (2022)
- N. Oku, S. Takemura, H. Onaka*, Y. Igarashi*. NMR characterization of streptogramin B and L-156,587, a non-synergistic pair of the streptogramin family antibiotic complexes produced inductively by a combined culture of Streptomyces albogriseolus and Tsukamurella pulmonis. MRC 60(2): 261-270 (2022)
2021
- T. Nagakubo, T. Yamamoto, S. Asamizu, M. Toyofuku, N. Nomura, H. Onaka*. Phage tail-like nanostructures affect microbial interactions between Streptomyces and fungi. Scientific Reports, 11(1), 20116 (2021)
- Y. Jiang, T. Matsumoto, T. Kuranaga, S. Lu, W. Wang, H. Onaka, H. Kakeya, Longicatenamides A-D, Two Diastereomeric Pairs of Cyclic Hexapeptides Produced by Combined-culture of Streptomyces sp. KUSC_F05 and Tsukamurella pulmonis TP-B0596. The Journal of Antibiotics, 74, 307–316 (2021)
- C. Pan, T. Kuranaga, X. Cao, T. Suzuki, N. Dohmae, N. Shinzato, H. Onaka, H. Kakeya, Amycolapeptins A and B, Cyclic Nonadepsipeptides Produced by Combined-culture of Amycolatopsis sp. and Tsukamurella pulmonis. The Journal of Organic Chemistry 86(2), 1843-1849 (2021)
2020
- Vinogradov AA*, Nagai E, Chang JS, Narumi K, Onaka H*, Goto Y*, Suga H*. Accurate Broadcasting of Substrate Fitness for Lactazole Biosynthetic Pathway from Reactivity-Profiling mRNA Display. Journal of the American Chemical Society, 142, 48, 20329–20334 (2020) doi: 10.1021/jacs.0c10374.
- A. A. Vinogradov, M. Shimomura, N. Kano, Y. Goto, H. Onaka, and H. Suga.* Promiscuous enzymes cooperate at the substrate level en route to lacatzole A. Journal of the American Chemical Society, 142, 32, 13886–13897 (2020)
- R. Kozakai, T. Ono, S. Hoshino, H. Takahashi, Y. Katsuyama, Y. Sugai, T. Ozaki, K. Teramoto, K.Teramoto, K. Tanaka, I. Abe, S. Asamizu, and H. Onaka*. Acyltransferase that catalyses the condensation of polyketide and peptide moieties of goadvionin hybrid lipopeptides. Nature Chemistry, 12: 869–877 (2020); doi: 10.1038/s41557-020-0508-2.
- A. A. Vinogradov, M. Shimomura, Y. Goto, T. Ozaki, S. Asamizu, Y. Sugai, H. Suga*, H. Onaka*. Minimal lactazole scaffold for in vitro thiopeptide bioengineering. Nature Communications, 2020; 11: 2272. doi: 10.1038/s41467-020-16145-4
- L. Vaario*, S. Asamizu, T. Sarjala, N. Matsushita, H. Onaka, Y. Xia, H. Kurokochi, S. Morinaga, J. Huang, S. Zhang, C. Lian. Bioactive properties of streptomyces may affect the dominance of Tricholoma matsutake in shiro. Symbiosis, 1-13 (2020)
2019
- Y Jiang, S Lu, G Hirai, T Kato, H Onaka, H Kakeya*. Enhancement of saccharothriolide production and discovery of a new metabolite, saccharothriolide C2, by combined-culture of Saccharothrix sp. and Tsukamurella pulmonis. Tetrahedron Letters, 60 (15), 1072-1074 (2019)
- R. Sugiyama, T. Nakatani, S. Nishimura, K. Takenaka, T. Ozaki, S. Asamizu, H. Onaka, H. Kakeya*. Chemical Interactions of Cryptic Actinomycete Metabolite 5‐Alkyl‐1, 2, 3, 4‐tetrahydroquinolines through Aggregate Formation. Angewandte Chemie International Edition, 131 (38), 13620-13625 (2019)
- Hoshino S, Onaka H, Abe I. Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. Journal of Industrial Microbiology & Biotechnology 46(3-4): 363-374 (2019)
- Ozaki T, Sugiyama R, Shimomura M, Nishimura S, Asamizu S, Katsuyama Y, Kakeya H, Onaka H*. Identification of the common biosynthetic gene cluster for both antimicrobial streptoaminals and antifungal 5-alkyl-1,2,3,4-tetrahydroquinolines. Org Biomol Chem. in press (2019) doi: 10.1039/c8ob02846j
2018
- Hoshino S, Onaka H, Abe I. Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. J Ind Microbiol Biotechnol. in press (2018) doi: 10.1007/s10295-018-2100-y
- Hoshino S, Ozeki M, Awakawa T, Morita H, Onaka H, Abe I. Catenulobactins A and B, Heterocyclic Peptides from Culturing Catenuloplanes sp. with a Mycolic Acid-Containing Bacterium. J Nat Prod. 81(9): 2106-2110 (2018) doi: 10.1021/acs.jnatprod.8b00261
- Hagihara R, Katsuyama Y, Sugai Y, Onaka H, Ohnishi Y. Novel desferrioxamine derivatives synthesized using the secondary metabolism-specific nitrous acid biosynthetic pathway in Streptomyces davawensis. J Antibiot (Tokyo). 71(11): 911-919 (2018) doi: 10.1038/s41429-018-0088-1.
- Hoshino S, Ozeki M, Wong CP, Zhang H, Hayashi F, Awakawa T, Morita H, Onaka H, Abe I. Mirilactams C-E, Novel Polycyclic Macrolactams Isolated from Combined-Culture of Actinosynnema mirum NBRC 14064 and Mycolic Acid-Containing Bacterium. Chem Pharm Bull (Tokyo). 66(6): 660-667 (2018) doi: 10.1248/cpb.c18-00143.
- Hoshino S, Wong CP, Ozeki M, Zhang H, Hayashi F, Awakawa T, Asamizu S, Onaka H, Abe I. Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium. J Antibiot (Tokyo). 71(7): 653-657 (2018) doi: 10.1038/s41429-018-0040-4.
2017
- Hoshino, S., Okada, M., Awakawa, T., Asamizu, S., Onaka, H., Abe, I*. Mycolic Acid Containing Bacterium Stimulates Tandem Cyclization of Polyene Macrolactam in a Lake Sediment Derived Rare Actinomycete. Org Lett 19, 4992-4995 (2017) doi:10.1021/acs.orglett.7b02508
- Onaka, H*. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J Antibiot (Tokyo) 70, 865-870 (2017) doi:10.1038/ja.2017.51
- T. Ozaki, K. Yamashita, Y. Goto, M. Shimomura, S. Hayashi, S. Asamizu, Y. Sugai, H. Ikeda, H. Suga*, and H. Onaka*. Dissection of goadsporin biosynthesis by in vitro reconstitution leading to designer analogs expressed in vivo. Nature communications 8, 14207 (2017) doi:10.1038/ncomms14207
2016
- Sugiyama R, Nishimura S, Ozaki T, Asamizu S, Onaka H, Kakeya H. Discovery and Total Synthesis of Streptoaminals: Antimicrobial [5,5]-Spirohemiaminals from the Combined-Culture of Streptomyces nigrescens and Tsukamurella pulmonis. Angew Chem Int Ed Engl. 55(35): 10278–10282 (2016) doi: 10.1002/anie.201604126.
- Du D, Katsuyama Y, Onaka H, Fujie M, Satoh N, Shin-Ya K, Ohnishi Y. Production of a Novel Amide-Containing Polyene by Activating a Cryptic Biosynthetic Gene Cluster in Streptomyces sp. MSC090213JE08. Chembiochem. 17(15) :1464-71. doi: 10.1002/cbic.201600167.
2015
- Ozaki T, Kurokawa Y, Hayashi S, Oku N, Asamizu S, Igarashi Y, Onaka H. Insights into the biosynthesis of dehydroalanines in goadsporin. ChemBioChem 2015 Dec 2. doi: 10.1002/cbic.201500541.
- Hoshino S, Okada M, Wakimoto T, Zhang H, Hayashi F, Onaka H, Abe I. Niizalactams A-C, Multicyclic Macrolactams Isolated from Combined Culture of Streptomyces with Mycolic Acid-Containing Bacterium. J Nat Prod 2015 Dec 1. in press
- Asamizu S, Ozaki T, Teramoto K, Satoh K, Onaka H. Killing of Mycolic Acid-Containing Bacteria Aborted Induction of Antibiotic Production by Streptomyces in Combined-Culture. PLoS One 2015 Nov 6;10(11):e0142372. doi: 10.1371/journal.pone.0142372.
- Hoshino S, Wakimoto T, Onaka H, Abe I., Chojalactones A-C, Cytotoxic Butanolides Isolated from Streptomyces sp. Cultivated with Mycolic Acid Containing Bacterium. Org Lett 2015 Mar 20;17(6):1501-4. doi: 10.1021/acs.orglett.5b00385.
- Sugiyama R, Nishimura S, Ozaki T, Asamizu S, Onaka H, Kakeya H. 5-Alkyl-1,2,3,4-tetrahydroquinolines, New Membrane-Interacting Lipophilic Metabolites Produced by Combined Culture of Streptomyces nigrescens and Tsukamurella pulmonis. Org Lett 2015 Apr 17;17(8):1918-21. doi: 10.1021/acs.orglett.5b00607.
- H. Onaka, T. Ozaki, Y. Mori, M. Izawa, S. Hayashi, and S. Asamizu. Mycolic acid-containing bacteria activate heterologous secondary metabolite expression in Streptomyces lividans. J Antibiot (Tokyo) 2015 Apr 1. doi: 10.1038/ja.2015.31.
2014
- Hoshino S, Zhang L, Awakawa T, Wakimoto T, Onaka H, Abe I. Arcyriaflavin E, a new cytotoxic indolocarbazole alkaloid isolated by combined-culture of mycolic acid-containing bacteria and Streptomyces cinnamoneus NBRC 13823. J Antibiot (Tokyo) 2014 Oct 22. doi: 10.1038/ja.2014.147.
- K.Haginaka, S. Asamizu, T. Ozaki, Y. Igarashi, T. Furumai, and H. Onaka*. Genetic approaches to generate hyper-producing strains of goadsporin: the relationships between productivity and gene duplication in secondary metabolite biosynthesis. Biosci Biotechnol Biochem 78(3): 394-399 (2014)
- R. Uchida, M. Nakai, S. Ohte, H. Onaka, T. Katagiri, and H. Tomoda*. 5-Prenyltryptophol, a new inhibitor of bone morphogenetic protein-induced alkaline phosphatase expression in myoblasts, produced by Streptomyces colinus subsp. albescens HEK608. J Antibiot (Tokyo) 2014 May 14. doi: 10.1038/ja.2014.44.
- S. Hayashi, T. Ozaki, S. Asamizu, H. Ikeda, S. Ōmura, N. Oku, Y. Igarashi, H. Tomoda, and H. Onaka*. Genome mining reveals a minimum gene set for the biosynthesis of 32-membered macrocyclic thiopeptides lactazoles. Chem Biol 21(5): 679-88 (2014)
2013
- Y. Kim, Y. In, T. Ishida, H. Onaka, and Y. Igarashi*. Biosynthetic origin of alchivemycin A, a new polyketide from Streptomyces and absolute configuration of alchivemycin B. Org Lett 15(14): 3514-3517 (2013)
- M. Kameya, H. Onaka, and Y. Asano*. Selective tryptophan determination using tryptophan oxidases involved in bis-indole antibiotic biosynthesis., Anal Biochem 438(2): 124-132 (2013)
- N. Koyama, M. Yotsumoto, H. Onaka. and H. Tomoda*. New structural scaffold 14-membered macrocyclic lactone ring for selective inhibitors of cell wall peptidoglycan biosynthesis in Staphylococcus aureus., J Antibiot (Tokyo) 66(5): 303-304 (2013)
2012
- Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian KD, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJ, Rebuffat S, Ross RP, Sahl HG, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, Smith L, Stein T, Süssmuth RD, Tagg JR, Tang GL, Truman AW, Vederas JC, Walsh CT, Walton JD, Wenzel SC, Willey JM, and van der Donk WA*. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30(1): 108-160 (2012)
- S. Asamizu, S. Hirano, H. Onaka*, H. Koshino, Y. Shiro, and S. Nagano*. Coupling Reaction of Indolepyruvic Acid in the Presence of StaD and Its Product: Implications for Biosynthesis of Indolocarbazole and Violacein. ChemBioChem 13(17): 2495-2500 (2012)
- K. Sakai, N. Koyama, T. Fukuda, Y. Mori, H. Onaka, and H. Tomoda*. Search method for inhibitors of Staphyloxanthin production by methicillin-resistant Staphylococcus aureus. Biol Pharm Bull 35(1): 48-53 (2012)